WP4 – Integrative genotype-phenotype data analysis
| WP4 | Integrative genotype-phenotype data analysis | WP Leader: Hubert Paush (ETH Zürich) | ETH Zürich, FBN, INRAE, ULIEGE, WR, AU, Luke, UEDIN |
|---|---|---|---|
| Task 4.1 | Association testing between whole-genome sequence variants and BovReg key traits | Task Leader: Hubert Pausch (ETH Zürich) | ETH Zürich, DPIV, AU, FBN, INRAE, LUKE, UALBERTA, UNILIM, WR |
| Task 4.2 | Effects of integration sites | Task Leader: Carole Charlier (ULIEGE) | ULIEGE, ETH Zürich |
| Task 4.3 | eQTL and mQTL studies | Task Leader: Christa Kühn (FBN) | FBN, DPIV, AU, ULIEGE, LUKE, UALBERTA |
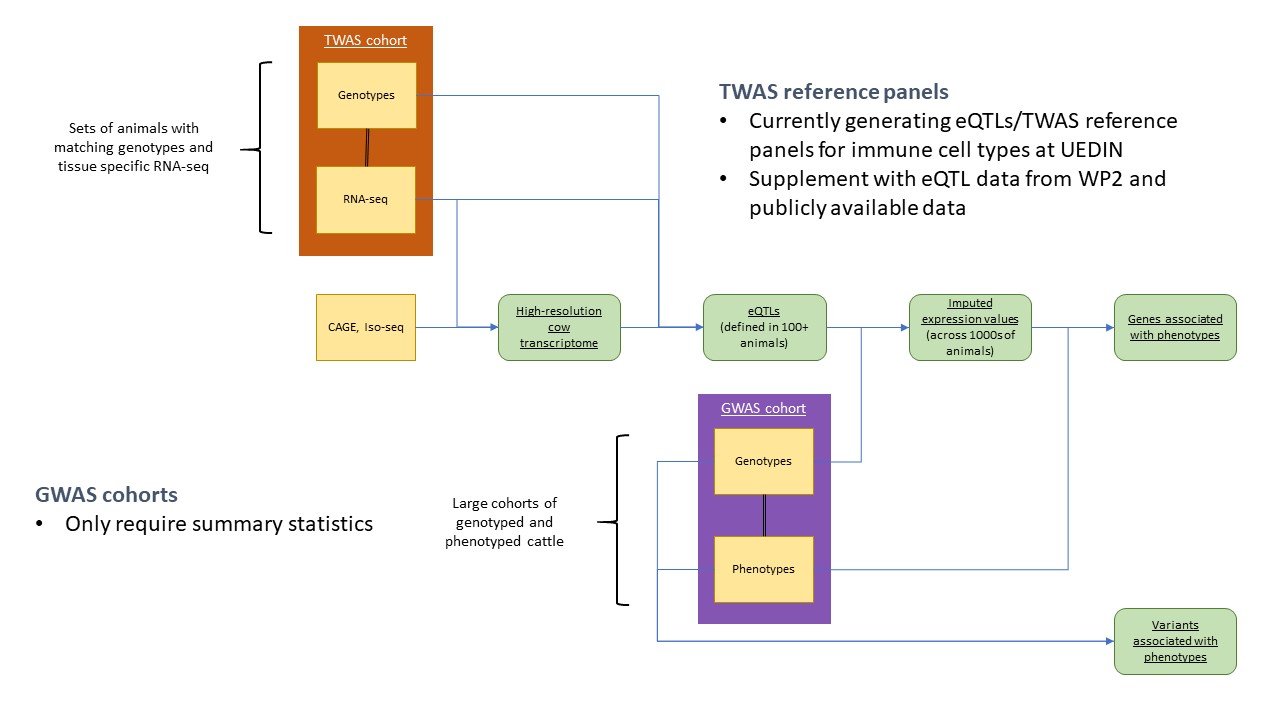
| Task 4.4 | Integrating Omics data in GWAS | Task Leader: Emily Clark (UEDIN) | UEDIN, ETH Zürich, FBN, LUKE |
WP4 objectives
Utilising the map of functionally active elements in the bovine genome (WP2, 5), WP4 will combine WGS, transcriptomic, metabolite and phenotypic data from beef and dairy cattle to pinpoint genetic variants underpinning variation in gene expression, intermediate phenotypes and traits related to biological efficiency, disease resistance and fertility.