WP5 – Epigenetics and environmental impact
| WP5 | Epigenetics and environmental impact | WP Leader: Jens Vanselow (FBN) | FBN, INRAE, ULIEGE, LUKE |
|---|---|---|---|
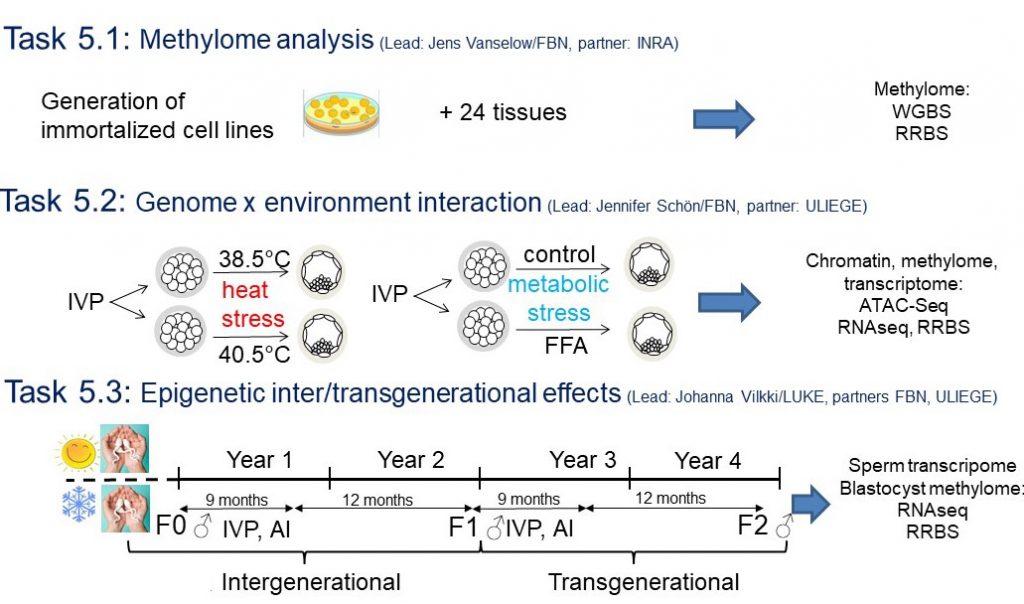
| Task 5.1 | Methylome analysis | Task Leader: Jens Vanselow (FBN) | FBN, INRAE |
| Task 5.2 | Genome x environment interaction | Task Leader: Jennifer Schön (FBN) | FBN, ULIEGE |
| Task 5.3 | Epigenetic inter/transgenerational effects | Task Leader: Johanna Vilkki (LUKE) | LUKE, FBN, ULIEGE |
WP5 objectives
Epigenetic marks (DNA methylation, chromatin structure) will be mapped genome-wide in the BovReg cell lines (WP1) and tissues and in cultured bovine embryos. This will contribute to the map of functionally active structural elements (WP2). In addition, epigenetic consequences of environmental stressors (heat or metabolic stress) will be mapped, and mechanisms of paternal inter- and transgenerational transmission of stressor-induced epigenetic changes will be described. This way, differential epigenetic imprints in the bovine genome will be identified. The data will be compared and superimposed with data from WP2, 4 and 6 to generate a comprehensive map of functional regulatory sequences within the bovine genome.